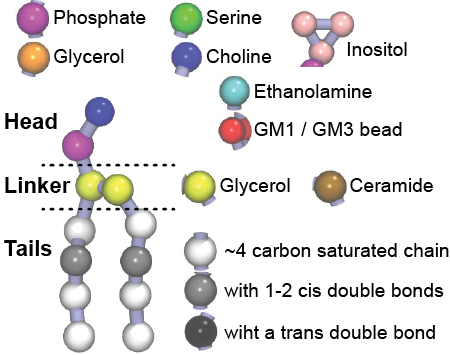
Improved Parameterization of Phosphatidylinositide Lipid Headgroups for the Martini 3 Coarse-Grain Force Field | Journal of Chemical Theory and Computation

Extending the Martini 3 Coarse-Grained Force Field to Carbohydrates | Journal of Chemical Theory and Computation

Evaluating Coarse-Grained MARTINI Force-Fields for Capturing the Ripple Phase of Lipid Membranes | bioRxiv

Overcoming the Limitations of the MARTINI Force Field in Simulations of Polysaccharides | Journal of Chemical Theory and Computation

Evaluating Coarse-Grained MARTINI Force-Fields for Capturing the Ripple Phase of Lipid Membranes | The Journal of Physical Chemistry B

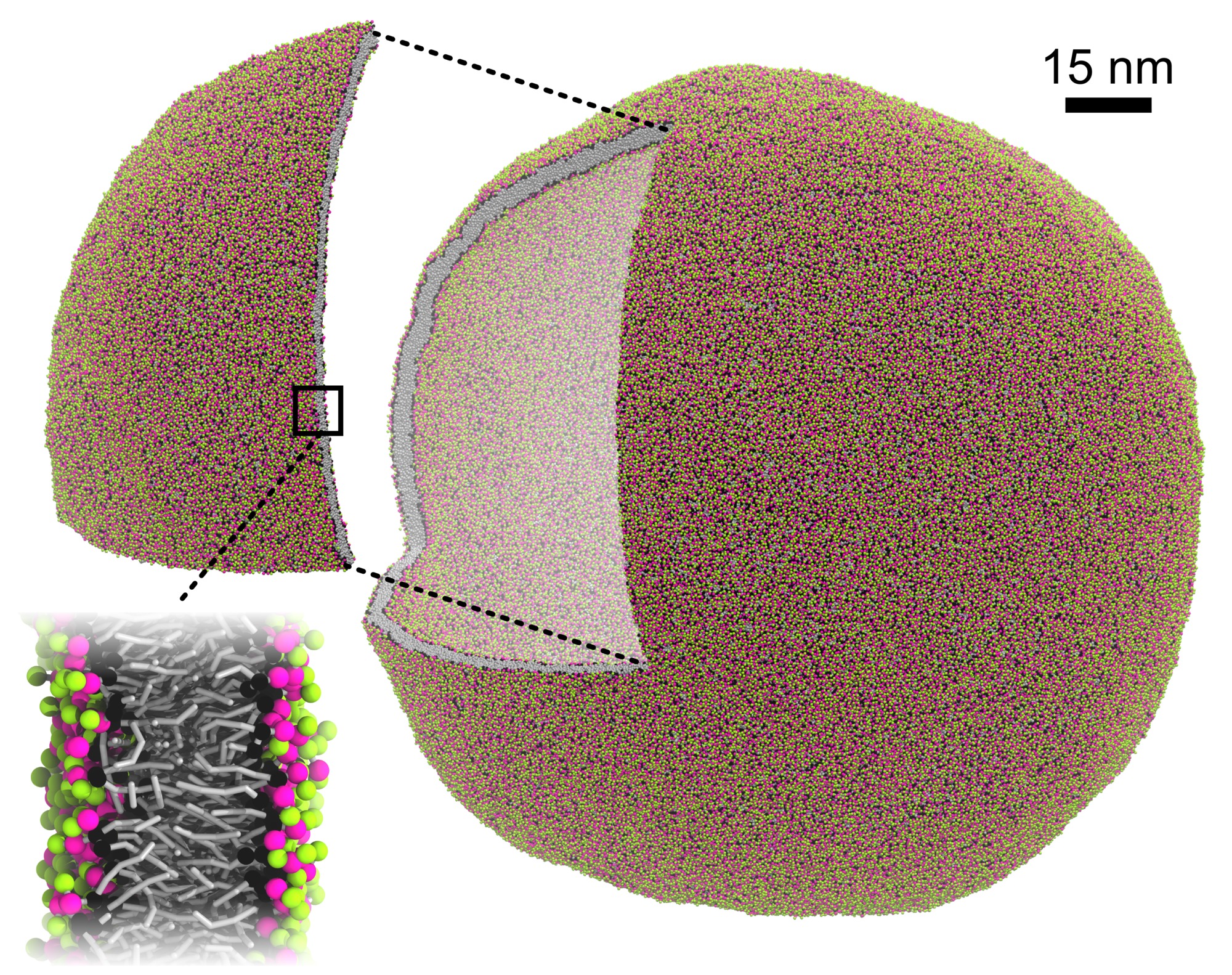
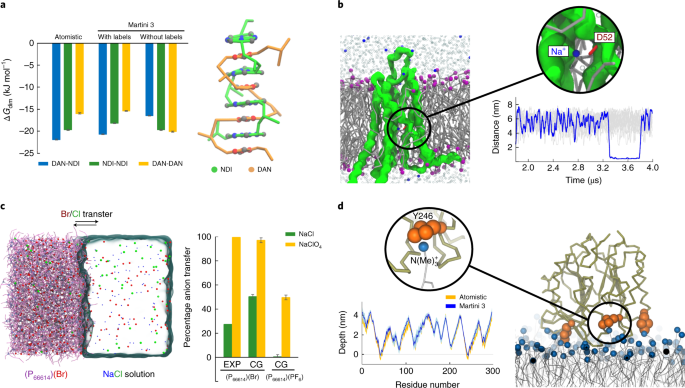
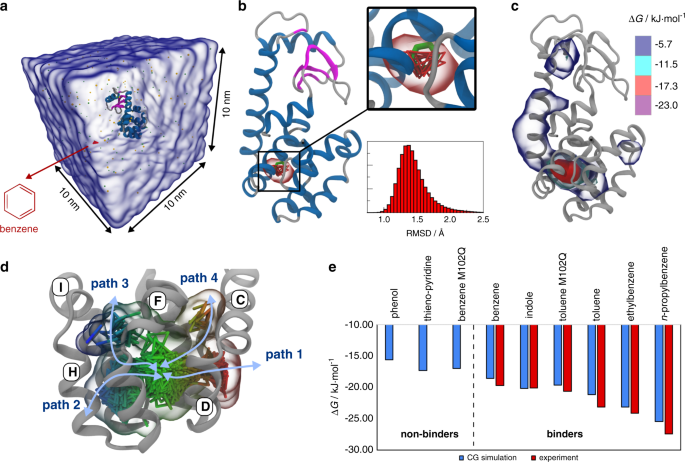
Two decades of Martini: Better beads, broader scope - Marrink - 2023 - WIREs Computational Molecular Science - Wiley Online Library

Swarm-CG: Automatic Parametrization of Bonded Terms in MARTINI-Based Coarse-Grained Models of Simple to Complex Molecules via Fuzzy Self-Tuning Particle Swarm Optimization | ACS Omega
Frontiers | Molecular Modeling for Nanomaterial–Biology Interactions: Opportunities, Challenges, and Perspectives
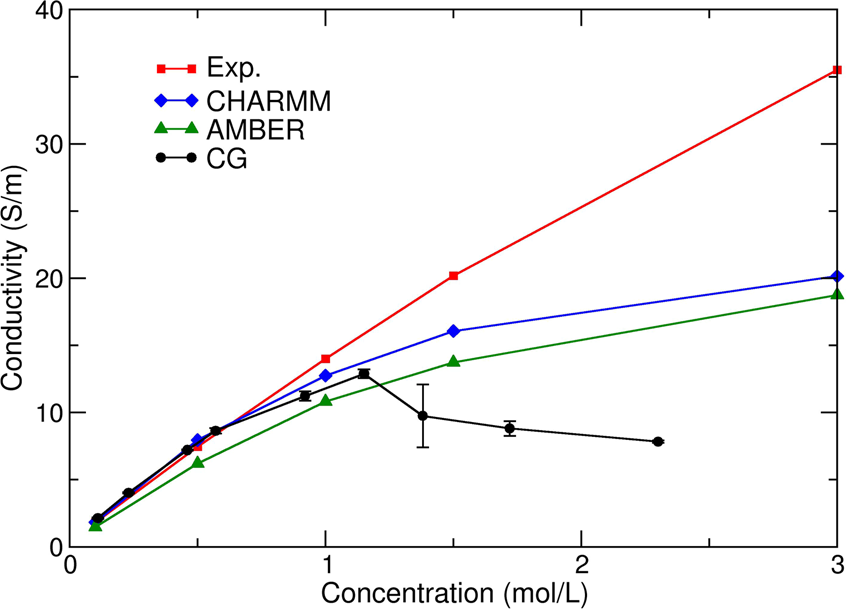
Ionic transport through a protein nanopore: a Coarse-Grained Molecular Dynamics Study | Scientific Reports

Two decades of Martini: Better beads, broader scope - Marrink - 2023 - WIREs Computational Molecular Science - Wiley Online Library
Lipid Clustering Correlates with Membrane Curvature as Revealed by Molecular Simulations of Complex Lipid Bilayers | PLOS Computational Biology

Swarm-CG: Automatic Parametrization of Bonded Terms in MARTINI-Based Coarse-Grained Models of Simple to Complex Molecules via Fuzzy Self-Tuning Particle Swarm Optimization | ACS Omega

Towards design of drugs and delivery systems with the Martini coarse-grained model | QRB Discovery | Cambridge Core