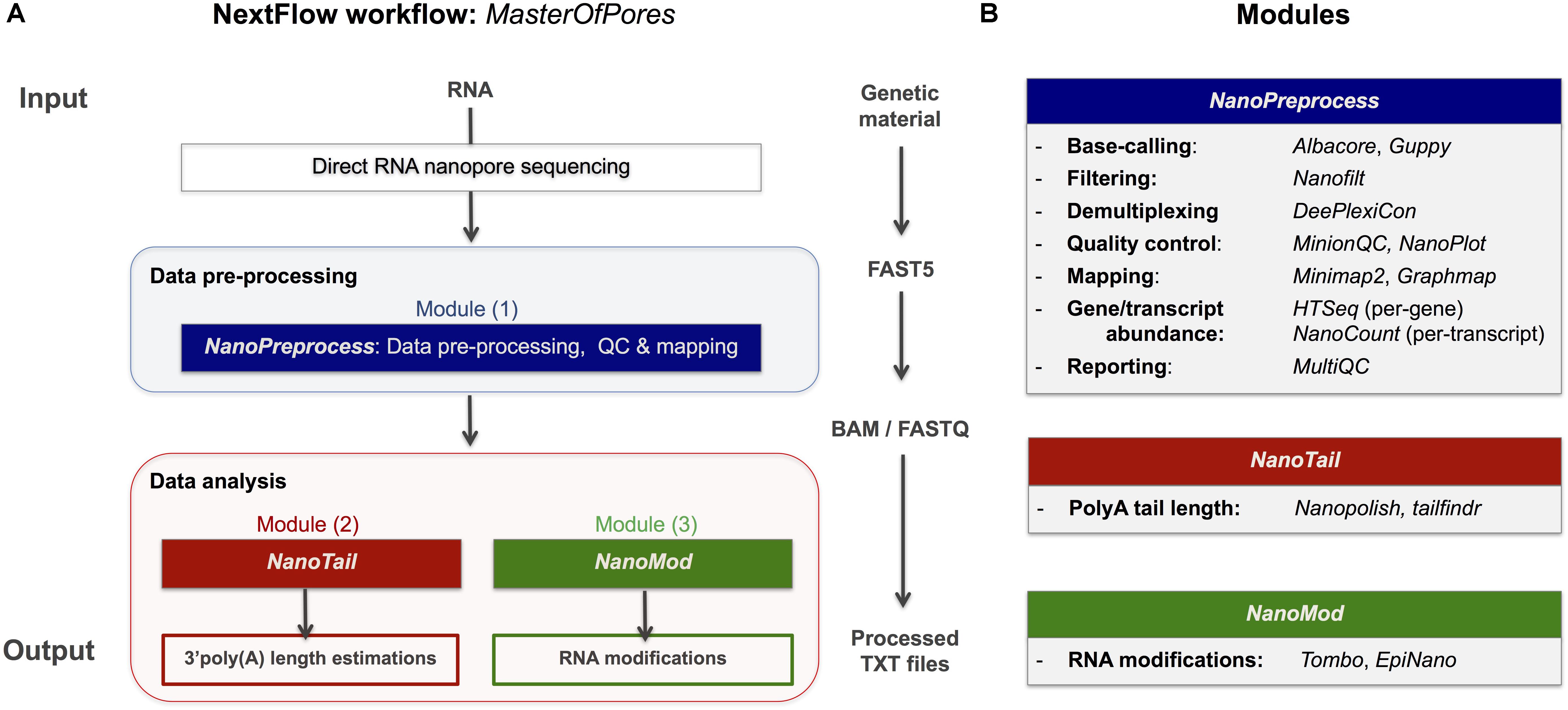
Frontiers | MasterOfPores: A Workflow for the Analysis of Oxford Nanopore Direct RNA Sequencing Datasets
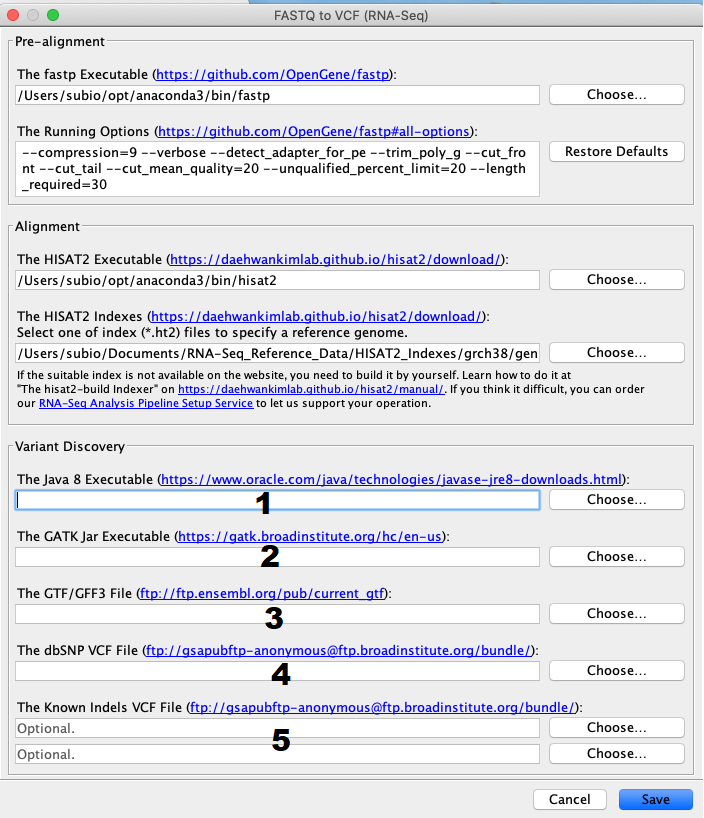
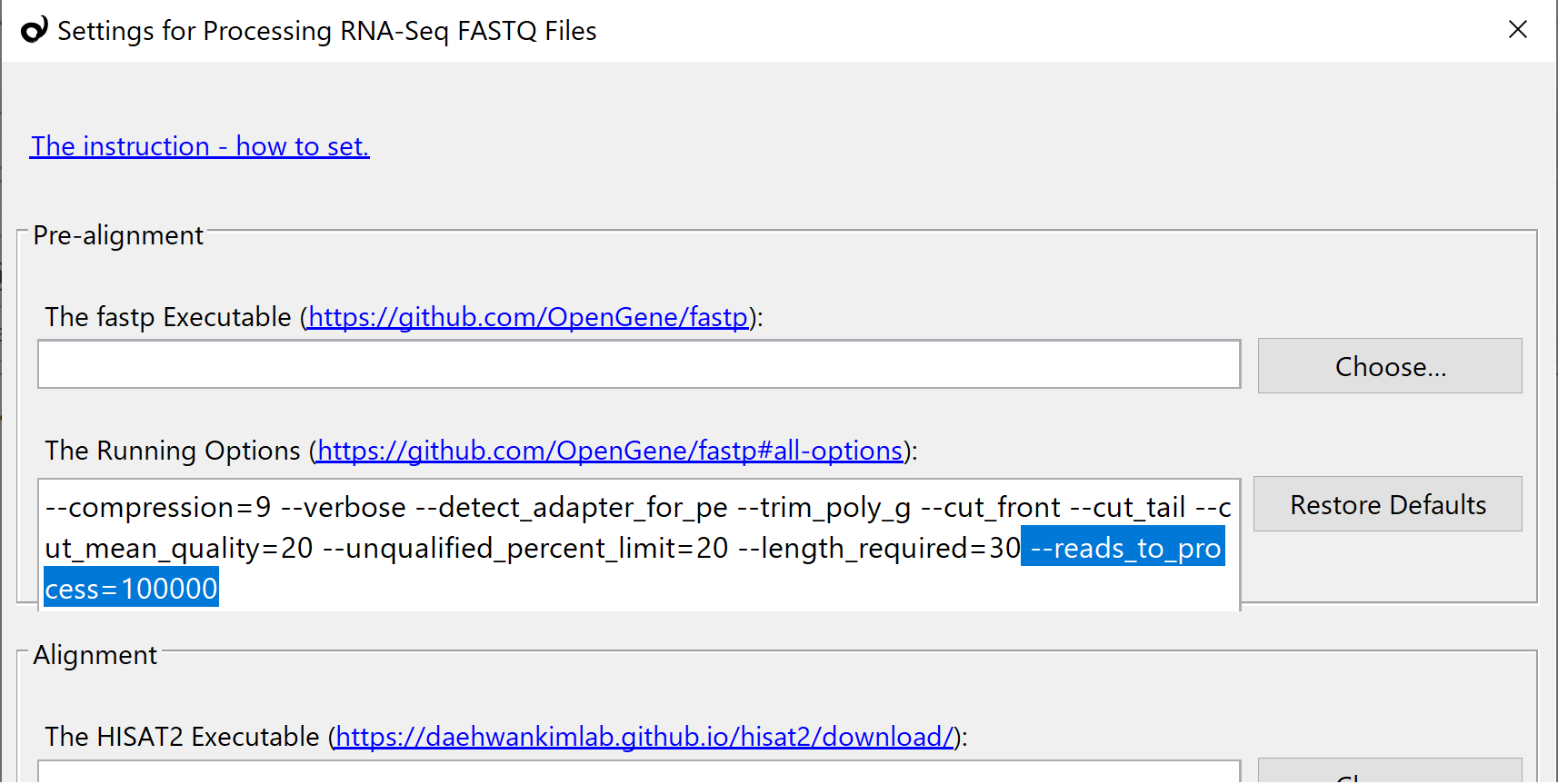
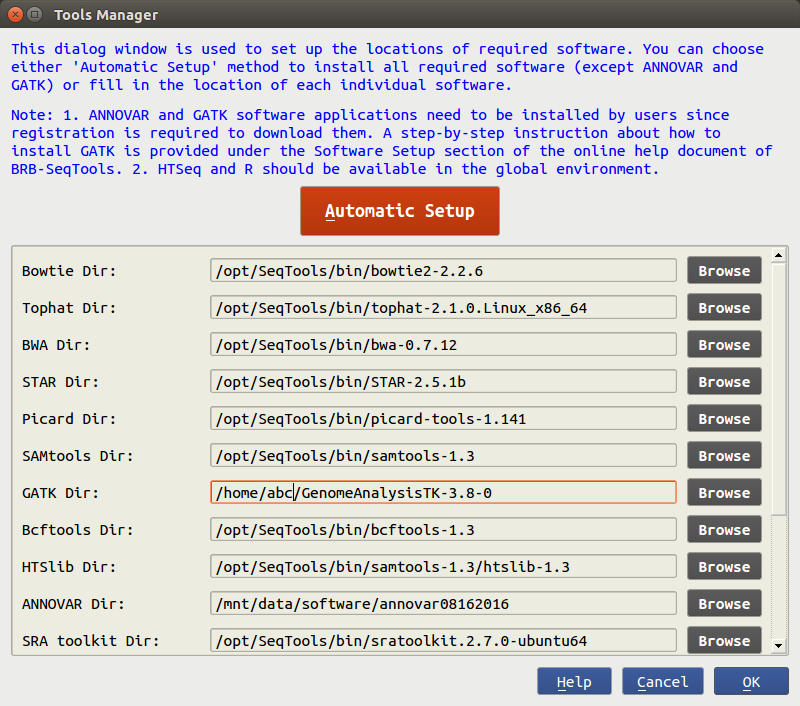
How to setup the environment for the pipeline of FASTQ to VCF (RNA-Seq) tool. (macOS version) | Subio

How long does it take to create single-end and pair-end data (fastq-dump) from SRAs? - usegalaxy.org support - Galaxy Community Help
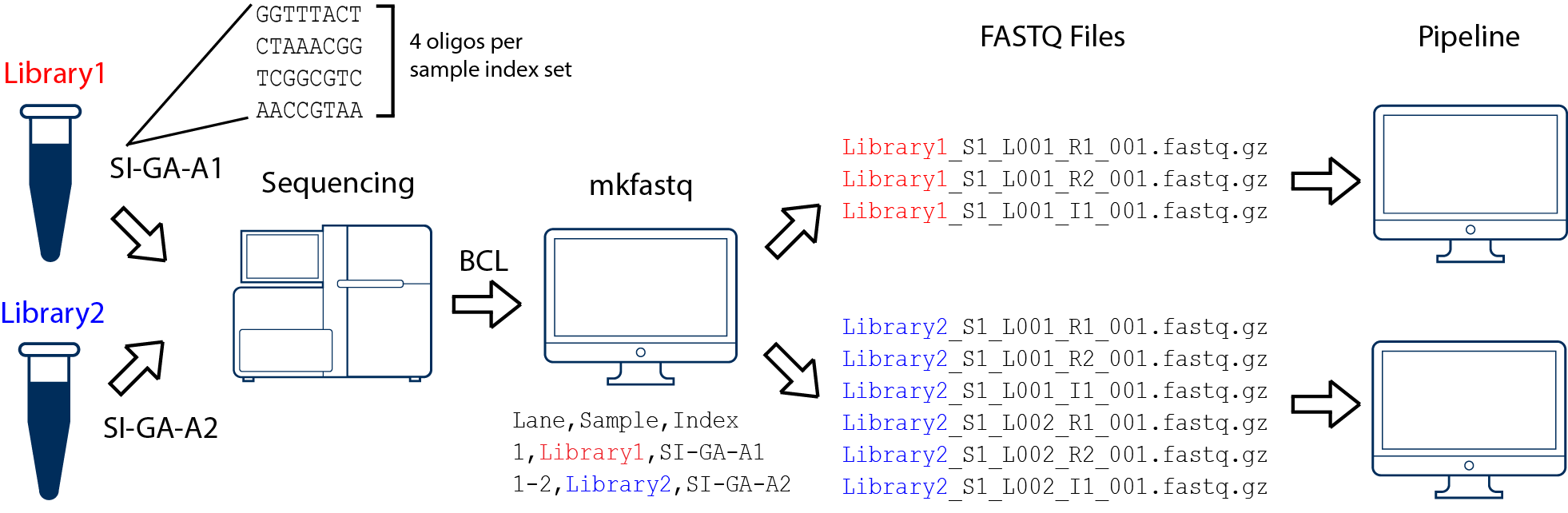

![2FAST2Q: a general-purpose sequence search and counting program for FASTQ files [PeerJ] 2FAST2Q: a general-purpose sequence search and counting program for FASTQ files [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2022/14041/1/fig-1-full.png)
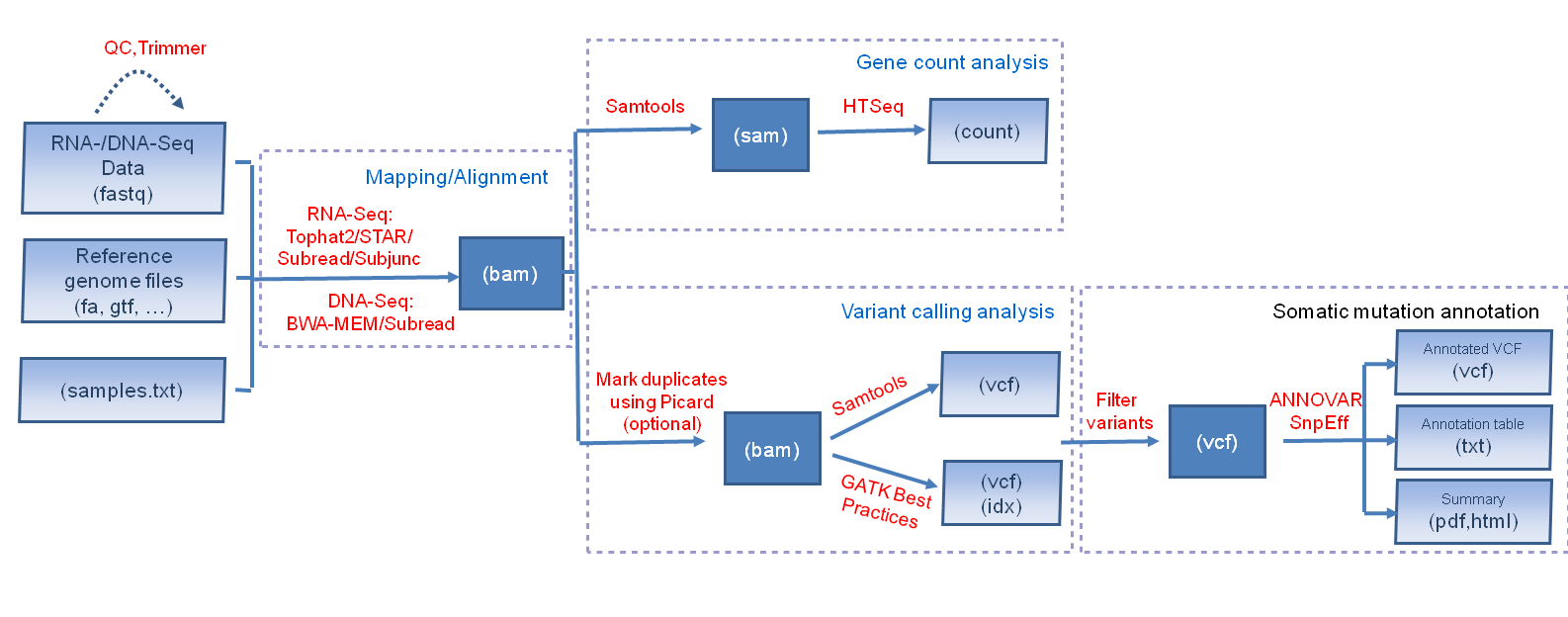




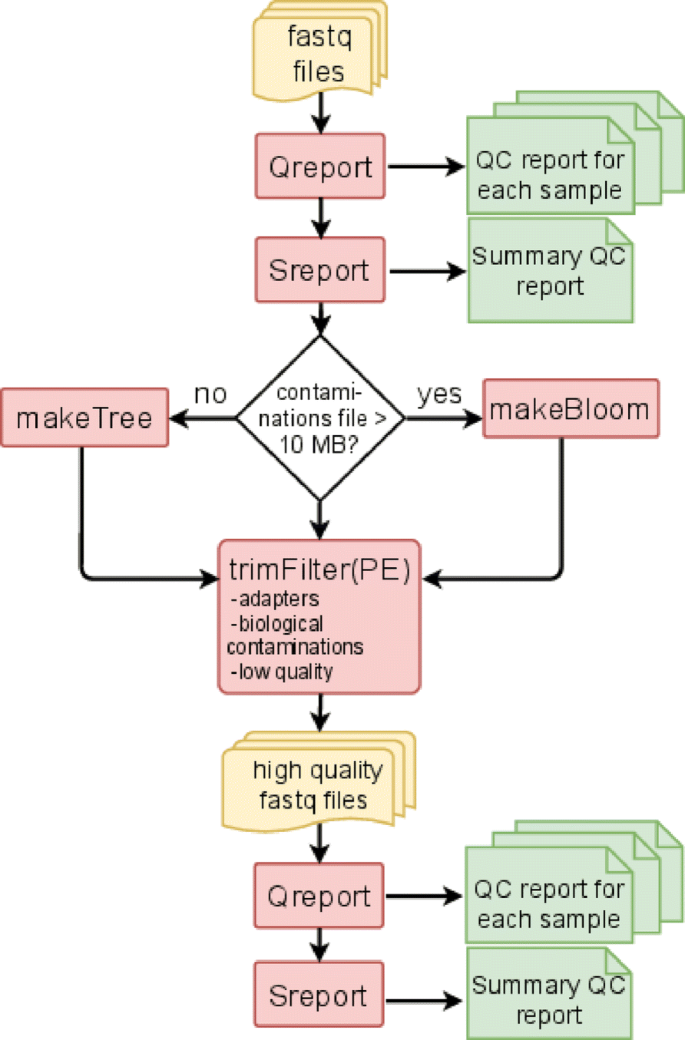



![2FAST2Q: a general-purpose sequence search and counting program for FASTQ files [PeerJ] 2FAST2Q: a general-purpose sequence search and counting program for FASTQ files [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2022/14041/1/fig-2-2x.jpg)